Projects
Contributing to the nf-core/proteinfold development – (NextFlow)

I am part of a team at the Structural Biology Facility - UNSW that is contributing to the international nf-core/proteinfold NextFlow pipeline, started at the Centre for Genomic Regulation in Barcelona. Proteinfold allows automatic high-performance protein folding from a simple samplesheet listing the target FASTA files. Proteinfold supports switching modes between multiple contemporary AI protein structure prediction programs, and generates an HTML report that visualises the protein structures. Since it’s written in NextFlow, it ensures reproducibility and can be run on either on-premises computing hardware or via cloud computing providers.
A main current of mine contribution to the pipeline is unifying the quality metrics from these AI models (structure confidence, interaction scores) into a common plaintext format that scientists can easily manipulate, rather than having to deserialise the various formats different programs have adopted and query particular keys. My aim is to amass these quality metrics into a summary MultiQC report Issue #439 that provides “at-a-glance” ranking and insight into structure prediction success of “bulk” protein folding (e.g. entire proteomes, oversampling replicates).
Folding entire proteomes with deep learning models – (AlphaFold)
I use high-performance computing systems to predict the folded structure of proteins from their amino acid sequence. By selecting efficient methods with acceptable accuracy for their application, I am able to “fold” the entire closed genome of organisms on modest high-performance compute servers. I then generate structured summaries and high quality rendered images, handing this molecular inventory back to biomolecular specialists who are able to identify key proteins in biochemical pathways.
Refactoring code for mass spectrometry reports – (R, GitHub)
I did some restructuring and refactoring of a codebase for a suite of mass spectrometry analysis reports, which had built up over time without consistent version control so had become divergent. Working alongside a talented Master’s student & R developer I migrated the set of core functions and code versions to an Enterprise GitHub repository and began identifying points of commonality between versions of plotting and statistical functions. This code rewrite significantly reduced the lines of code to maintain, made function naming and operation clearer, allowed easy addition and modification to analysis workflows, and enabled easy install from RStudio with install_github().
Structure-Activity Relationships for Carbonyl Photolysis – (PhD)
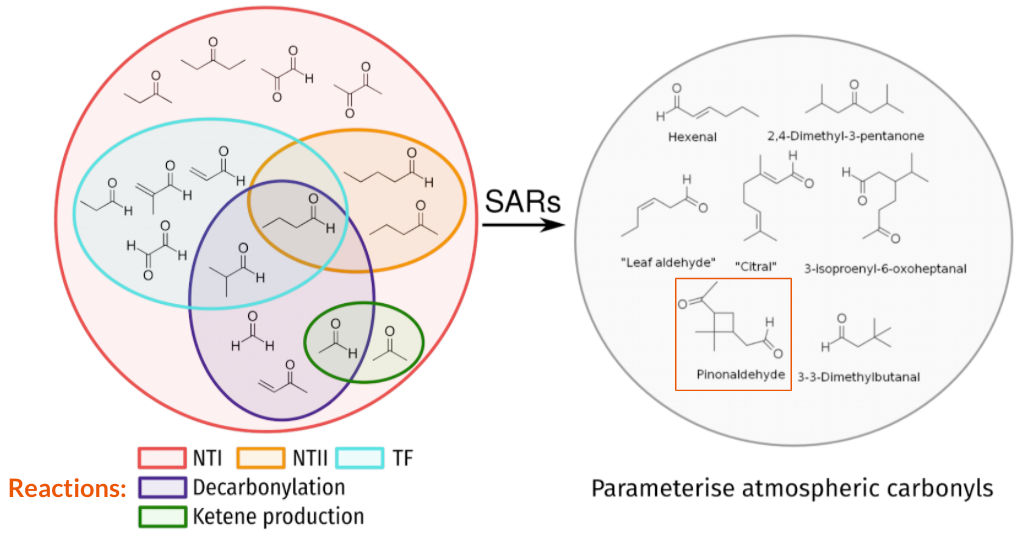
The photochemistry of diverse small carbonyls could be used to predict the behaviour of atmospherically important molecules.
My PhD thesis was on the development of a set of rules that define the relationships between the molecular structure of a carbonyl and how it reacts following UV light absorption. Carbonyls are central to the atmosphere, they are one of the few atmospheric molecules that absorb UV, the radicals they create from UV photolysis drive chemical cycles, and the C=O functional group is formed abundantly when carbon-containing molecules oxidise in the atmosphere. Atmospheric chemistry models manage the simulation of thousands of atmospheric molecules by using structure-activity relationships (SARs), but no SARs had been developed for carbonyl photolysis. Instead, cruder “surrogate” approximations are employed based upon experimental data for roughly a dozen molecules.
My PhD thesis developed SARs from comprehensive reaction threshold calculations on 38 representative carbonyls, across all relevant electronic states. My thesis is an attempt to derive more general carbonyl photolysis SARs to cover a wider range of molecules and reaction types. The dominant Norrish Type I photolysis reaction is covered in a paper in JPCA, and energetically accessible photo-initiated ground state reactions are covered in a paper in ACP. Preprints on the topics of other reactions and protocols are available on ChemRxiv
Revising Neighbour-Exclusion in DNA Intercalators – (Honours)
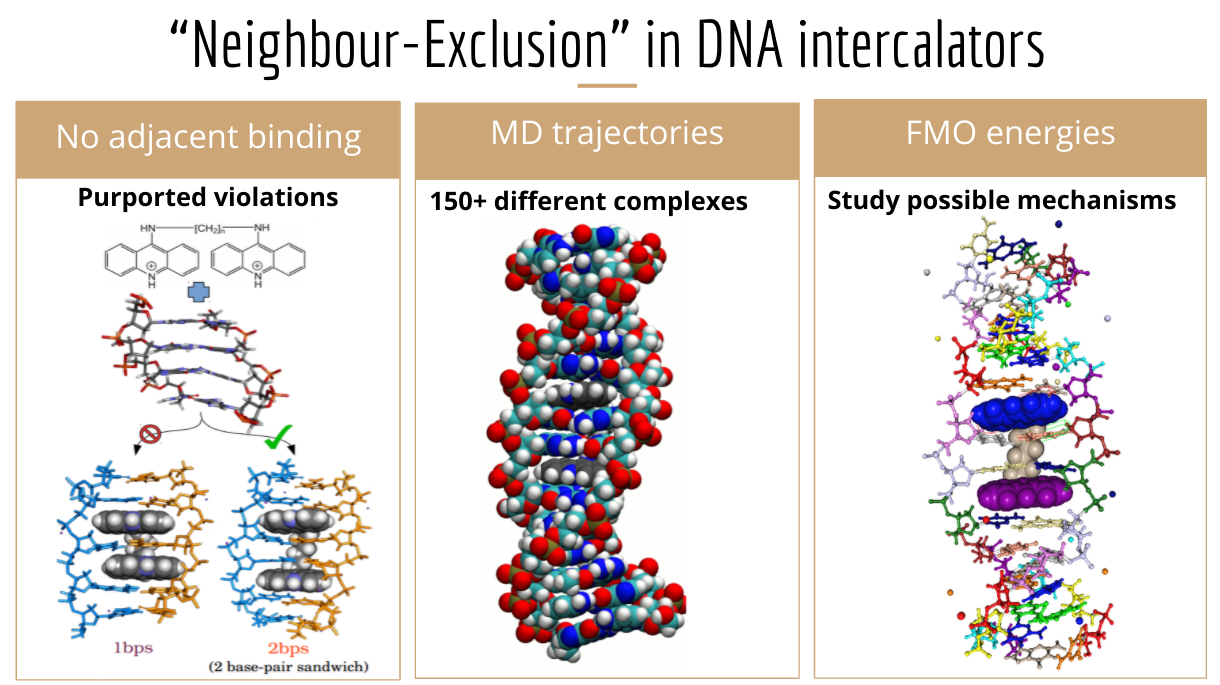
My simulations revise previous literature by indicating diacridines can insert into DNA without violating neighbour-exclusion.
I studied the insertion of diacridines (linked, flat aromatic molecules) into DNA for my Honours work, for which I received the top mark and Angyal Prize (Best performance in Honours Chemistry). I used molecular dynamics to study the structural feasibility of various binding modes, analysing the insertion into over hundred different structures, where the A,T,C,G sequence at the binding site, the diacridine linker chain length, and bound DNA groove were varied. These dynamics trajectories showed that insertion of both acridine ends was viable and in agreement with empirical measurements. I also performed fragment-based quantum chemical and free energy calculations to attempt to determine the basis of the neighbour-exclusion principle, i.e. insertion cannot occur at adjacent base-pairs, whose physical origins are still unclear. This work is detailed in a paper in Biopolymers.